Genome Editing: What does it mean to be human?
In 2015, the National Academies of Sciences, the US National Academy of Medicine, the Chinese Academy of Sciences, and the UK Royal Society hosted the International Summit on Human Gene Editing to discuss the ethics, laws, and future directions of genome editing, and its place in our society. These techniques have the power to add, replace, or remove specific genes from individuals, which can cure diseases or change the very nature of what it means to be human.
““[Genome editing] has the potential to cure rare diseases with a direct genetic cause.””
Genome editing is a rapidly expanding field of research that holds great promise for the medical sciences, which has the potential to cure rare diseases with a direct genetic cause. Some genome editing technologies that have been around for a while include homing endonucleases,1 transcription activator-like effector nucleases (TALENs),2 and zinc finger nucleases.3
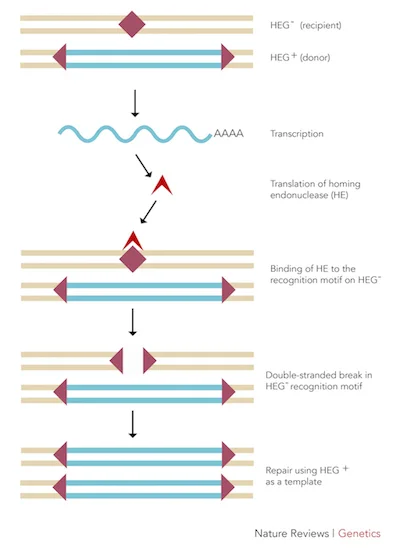
Figure 1: The mechanism by which homing endonucleases function.
Homing Endonucleases
Homing endonucleases are nucleases (enzymes that cleave nucleic acids, DNA and RNA, via hydrolytic cleavage of phosphodiester linkages) that cut DNA at a specific location and insert their own gene into that site.1 Once the homing endonuclease is synthesized, it locates the homologous chromosome that is paired with the chromosome that contains the endonuclease’s gene. It then identifies the area on the homologous pair that mirrors its gene locus and excises it out. Endogenous DNA repair mechanisms then fix the double-stranded breaks caused by the homing endonuclease by using the original chromosome (which contains the homing endonuclease gene) as a template. The homing endonuclease gene is then added into the homologous pair (Figure 1).1
TALENs
Transcription activator-like effector nucleases (TALENs) are essentially made up of the TAL effector domain, which binds to specific DNA sequences, and the DNA cleavage domain (e.g. FokI), which cuts the DNA.2 The TAL effector domain is derived from Xanthomonas proteins,2 which have a very simple correspondence between amino acid sequences and the nucleotides that they bind. This simplicity means that we can adapt these amino acid sequences and string them into a TAL effector that binds to any specific DNA sequence. In order to effectively create a cleavage site, two TALENs are needed to create a double-stranded break (DSB). The two TAL effectors must be engineered to bind to sites upstream and downstream of the desired cleavage site, respectively. This cleavage will become the site in which genes can be removed, modified, or added.2
Zinc Finger Nucleases
Another genome editing technology that is highly similar to TALENs is zinc finger nucleases, which are also nucleases that contain two unique domains: the FokI nuclease domain and a customizable DNA-binding domain that consists of 3-6 zinc fingers, protein motifs that require the presence of one or more zinc ions to stabilize them.3 Each zinc finger can bind to three nucleotides in the target DNA.3 By using multiple zinc fingers, each with a known nucleotide sequence specificity, it is possible to create a highly specific DNA-binding domain that binds to a target DNA sequence. Zinc finger domains are generally created by testing the specificity of randomized zinc fingers to a target sequence and picking the desirable ones.
However, the topic that spurred the International Summit was the clustered regularly interspaced short palindromic repeats (CRISPR) technology, which has multiple advantages over homing endonucleases, TALENs, and zinc finger nucleases such as high specificity for desired DNA sequences, effective selection of target sites, high efficiency, and simple design and construction.
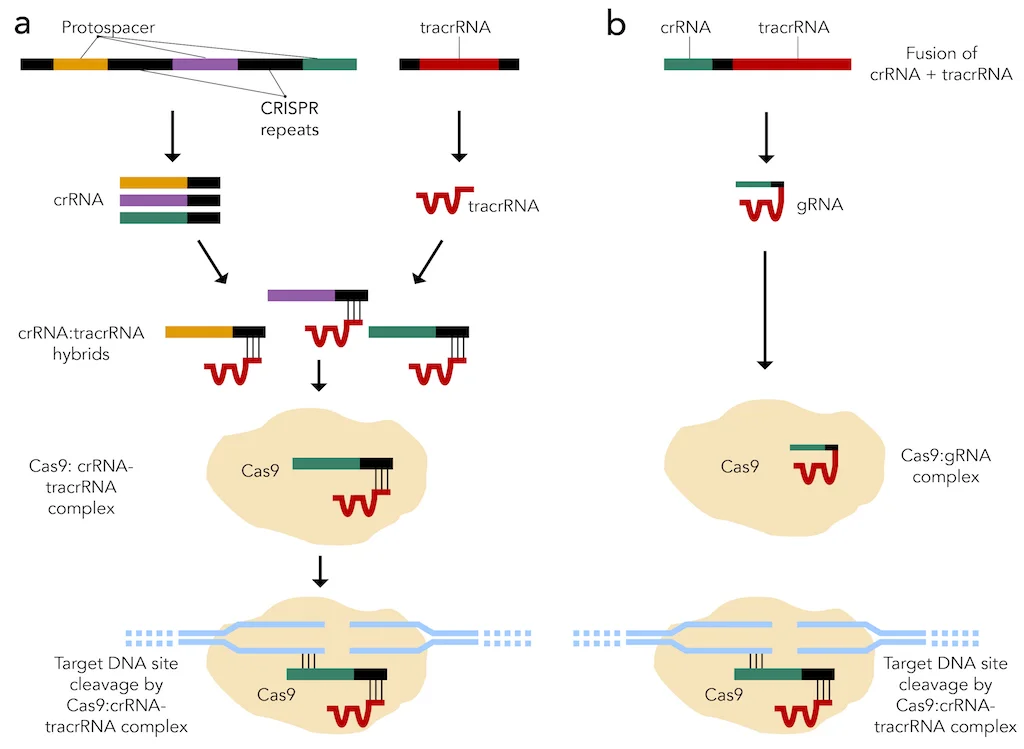
Figure 2: CRISPR-Cas9 systems that are (a) present in nature and (b) created synthetically.
CRISPR
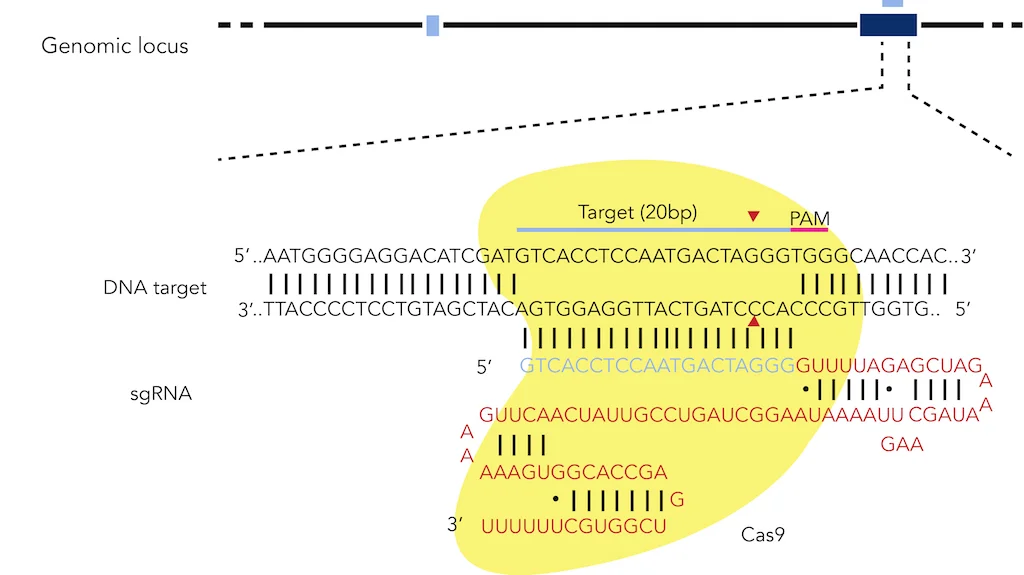
The CRISPR system is found naturally in bacteria and involves a CRISPR-associated 9 (Cas9) endonuclease used by small CRISPR RNAs (crRNAs) to cleave nearly any genomic locus. More specifically, these crRNAs bind to trans-activating crRNAs (tracrRNAs) to form hybrid RNA duplexes, which associate with the Cas9 endonuclease. This RNA-protein complex then goes to the target DNA sequence, where a region of the crRNA that is complementary to a portion of the target sequence, called the protospacer, binds to the target sequence (Figure 2a).5,6 Alternatively, scientists can fuse crRNA and tracrRNA together to yield a single guide RNA (sgRNA), which function in the same way as crRNA-tracrRNA duplexes (Figure 2b). A crucial element of the target DNA sequence is the protospacer adjacent motif (PAM) that immediately follows the portion of the target sequence that the protospacer recognizes for binding. For example, the highly conserved PAM for Streptococcus pyogenes (S. pyogenes) is the “NGG” sequence, where “N” stands for any nucleotide (Figure 3).4 After protospacer binding and PAM recognition, a double-stranded break (DSB) is introduced to the target DNA sequence by Cas9. Specific point mutations in either the protospacer on crRNAs or PAM on the target DNA sequence can disrupt CRISPR-mediated cleavage of the target sequence.6
Figure 3: CRISPR targeting of the human EXM1 locus with the Cas9 endonuclease from S. pyogenes.
Once Cas9 creates a double-stranded break (DSB) in the target DNA sequence, this locus undergoes one of two major DNA repair mechanisms. The first mechanism is non-homologous end joining (NHEJ), which re-ligates the separated DNA strands without the use of a DNA template. This repair pathway can produce mutations in the form of insertions and deletions (indels) at the DSB site, which may lead to frameshifts or premature stop codons within coding regions of the target sequence. The second mechanism is homology-directed repair (HDR), which requires the use of a DNA template to introduce small-scale mutations at the DSB site. The DNA templates incorporated in HDR have regions of sequence similarity to the target DNA sequence flanking a region containing the desired mutations (Figure 4). The CRISPR system has been successful in editing portions of bacterial genomes such as the srtA and bgaA loci of Streptococcus pneumoniae (S. pneumoniae) and the rpsL locus of Escherichia coli (E. coli),6 and sections of eukaryotic genomes such as the Th locus of mice and the EMX1 and PVALB loci of humans.5
Figure 4: Genome editing through using canonical DSB repair pathways.
Despite multiple advantages of the CRISPR system, a major disadvantage of this method of genome editing is the increased risk of off-target cleavage by the sgRNA-Cas9 complex on the target genome because some target DNA sequences that are similar to one another can be recognized by the same protospacer sequence. This risk is highlighted by the study of human CCR5 and HBB genes, which led to the generation of unwanted indels and point mutations.7 In light of this, researchers must consider genomic target sites that can be cleaved with high specificity.
““There is considerable hope for the widespread use of CRISPR to eradicate disease genotypes and phenotypes.””
CRISPR is fast becoming an invaluable asset in biology, especially in the correction of mutations attributed to genetic diseases. In the past couple of years alone, researchers used the CRISPR system in mouse models and cultured human cells to restore wild-type genes implicated in diseases such as cystic fibrosis (CF),8 dominant cataract disorder,9 Duchenne muscular dystrophy (DMD),10 and hereditary tyrosinemia type I (HTI).11 Therefore, there is considerable hope for the widespread use of CRISPR to eradicate disease genotypes and phenotypes, which are interlaced with many ethical implications that the International Summit sought to discuss.
Works Cited:
1. Stoddard BL. Homing endonuclease structure and function. Q Rev Biophys. 2005;38(01):49-95.
2. Bogdanove AJ, Voytas DF. TAL effectors: Customizable proteins for DNA targeting. Science. 2011;333(6051):1843-1846. doi: 10.1126/science.1204094.
3. Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nature Reviews Genetics. 2010;11(9):636-646.
4. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8(11):2281-2308.
5. Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819-823. doi: 10.1126/science.1231143.
6. Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-cas systems. Nat Biotechnol. 2013;31(3):233-239.
7. Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41(20):9584-9592. doi: 10.1093/nar/gkt714.
8. Schwank G, Koo B, Sasselli V, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell stem cell. 2013;13(6):653-658.
9. Wu Y, Liang D, Wang Y, et al. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell stem cell. 2013;13(6):659-662.
10. Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345(6201):1184-1188. doi: 10.1126/science.1254445.
11. Yin H, Xue W, Chen S, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014.
Cite This Article:
Chan G., Zheng K., Ho J. Genome Editing: What does it mean to be human? Illustrated by B. Sharma. Rare Disease Review. July 2017. DOI:10.13140/RG.2.2.16868.58248.